Introduction to pattern Matching
What is Pattern Matching?
It's the process of identifying specific sequences of characters or elements within a larger structure like text, data, or images. Think of it like finding a specific word in a sentence or a sequence of symbols or values, within a larger sequence or text.
Basic Concepts Pattern Matching
- Pattern: A pattern is a sequence of characters, symbols, or other data that forms a search criterion. In text processing, a pattern could be a string of characters.
- Text: The text (or string) is the sequence where the pattern is searched for.
- Match: A match occurs if the pattern is found within the text. The goal of pattern matching is to find all instances where this occurs or to determine whether the pattern exists in the text.
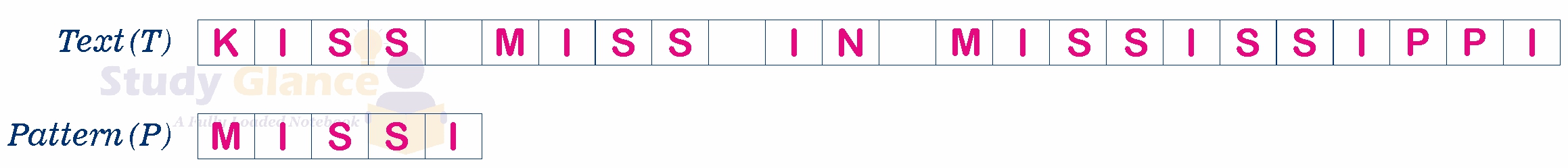
Common Algorithms of Pattern Matching
Brute Force Pattern Matching Algorithm
Checks for the pattern at every possible position in the text. For each position, it compares the pattern with the corresponding substring in the text. It is Effective for small texts or patterns, but inefficient for large texts.
Knuth-Morris-Pratt(KMP)
Optimizes the naive approach by avoiding redundant comparisons. It pre-processes the pattern to determine the longest prefix which is also a suffix, allowing the search to skip some comparisons. It is Suitable for applications where the same pattern is searched repeatedly in multiple texts.
Boyer-Moore
Works by comparing the pattern to the text from right to left. It uses two heuristics, the bad character rule and the good suffix rule, to skip sections of the text, offering potentially sub-linear time complexity. It is Highly efficient for large texts and is considered one of the fastest single-pattern matching algorithms.
Rabin-Karp
Uses hashing to find any set of pattern occurrences. It hashes the pattern and text's substrings of the same length and then compares these hashes. If the hashes match, it checks for a direct match. It is Useful in plagiarism detection or searching for multiple patterns simultaneously.
Finite Automata
Constructs a state machine based on the pattern. The text is then processed character by character, transitioning between states of the automaton. It is Effective when the same pattern is matched against many texts, as the automaton needs to be constructed only once.
Aho-Corasick Pattern Matching Algorithm
A more complex algorithm used for finding all occurrences of any of a finite number of patterns within the text. It constructs a trie of patterns and then a state machine from the trie. Ideal for matching a large number of patterns simultaneously, like in virus scanning or "grep" utilities.
Next Topic :Brute force Pattern Matching